Data Nutrition Label
We have a Data Nutrition Label (DNL) Mockup
Context
Data Nutrition Label (DNL) is a way to communicate the contents and quality of data to the client (user or front-end)
It is expected that the DNL will be generated and stored according to the Pipeline Storage documentation.
As this data structure is designed, we can then standardize what needs to be outputted from the pipeline and how the API will reference those outputs.
Standards across ALL datasets
This diagram is based on all the sections below:
Python Pydantic class definitions
ABOUT
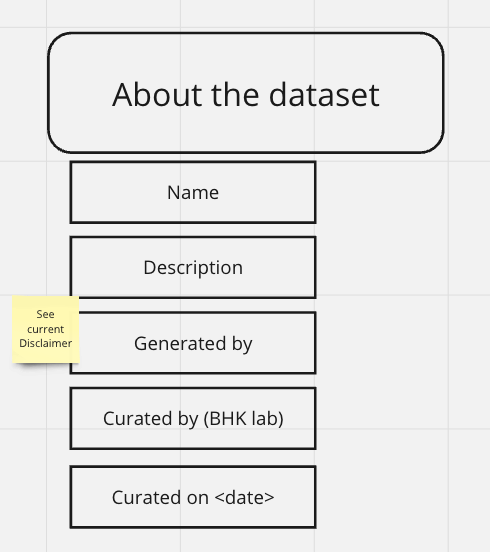
fld | value | source | default |
|---|---|---|---|
name | name of pipeline used across databases | API/client | NA |
description | outputted from the pipeline metadata file | Pipeline Output | NA |
Disclaimer | some disclaimer indicating usage? see disclaimer below | Pipeline Output | NA |
Curated By | Group that curated the pipeline | API/client | BHK Lab |
Curated On | Date of curation should be outputted as well | Pipeline Output | NA |
Disclaimer Data Usage Policy:The gCSI data were generated and shared by Genentech as part of the Genentech Cell Line Screening Initiative (Eva Lin, Yihong Yu, Scott Martin, Department of Discovery Oncology). The Haibe-Kains Lab has reprocessed and re-annotated the data to maximize overlap with other pharmacogenomic datasets.
Technical Information
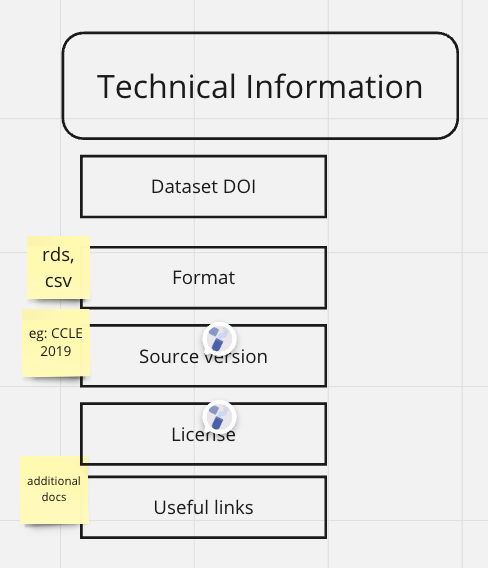
fld | value | source | default |
|---|---|---|---|
Dataset DOI | Zenodo DOI of the dataset | API/server | NA |
Data Type | Type of data (e.g | client/Pipeline Output | NA |
Data Format | Format of data (e.g | client/Pipeline Output | NA |
License | License of the data (e.g | github repo/pipeline output/client | Creative Commons Attribution 4.0 license |
Pipeline & Release Notes
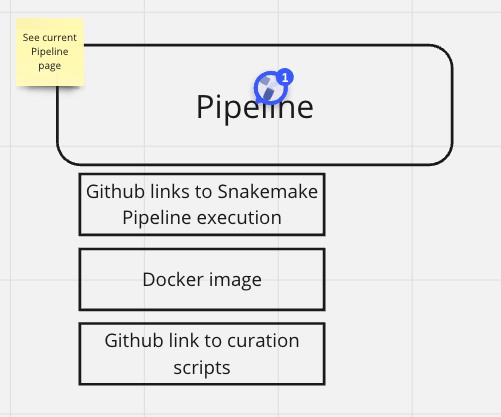

fld | value | source | default |
|---|---|---|---|
GitHub Repo | URL of the GitHub repository where the pipeline is stored | API/client | NA |
Release Notes | Notes on the pipeline release | repo/API/client | NA |
Data Quality
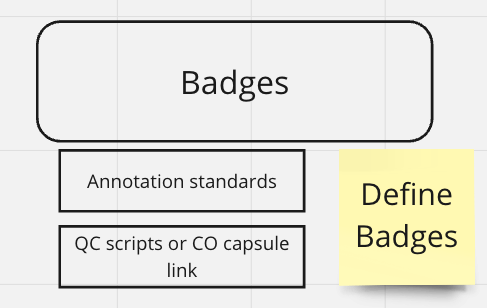
fld | value | source | default |
|---|---|---|---|
Annotation standards | ???? | ???? | ???? |
Quality Control | Link to an HTML file with the quality control metrics | ???? | ???? |